Crosswell Tomography Inversion 2D
12D Linear Inversion of Crosswell Tomography Data¶
import numpy as np
from geoscilabs.inversion.TomographyInversion import TomographyInversionApp
import matplotlib.pyplot as plt
from ipywidgets import interact, IntSlider
%matplotlib inline2Purpose¶
From the Linear inversion notebook (1D), we learned important apsects about the linear inversion. However, real world geophysical inverse problem are not often 1D, but multidimensional (2D or 3D), and this extension of dimension allows us to put more apriori (or geologic) information through the regularization term. In this notebook, we explore these multidimensional aspects of the linear inversion by using 2D traveltime croswell tomography example.
2.1Outline¶
This notebook includes three steps:
- Step 1: Generate a velocity model
- Step 2: Simulate traveltime data and add noise
- Step 3: Run inversion
2.2Step 1: Generate a velocity model¶
Here we set up a velocity model using a following app. Controlling parameters of the app are:
set mesh: use active only when you want to change the 2D meshadd block: use active when you want to add block (if not stay inactive)model type: background or blockshow grid?: show grid of the meshv0: velocity of the backgroundv1: velocity of the blockxc: x center of the blockyc: y center of the blockdx: width of the blockdz: thickness of the blocknx: # of cells in x-direction (this is only active whenset_mesh=active)ny: # of cells in y-direction (this is only active whenset_mesh=active)
Re-running the following cell will reset the app to the default parameters.
2.2.1Changing # of cells in - or - direction¶
Adjustable parameters: set mesh, nx, ny
The 2D domain is fixed at 200m × 400m. The number of cells in each direction can be changed which alters the size of cells in each direction. To change either nx or ny:
- Set
set mesh=active. This will change the view to the background modelv0. - Adjust
nxand ornyusing the sliders. The size of the cells is diplayed above the model. - When finished adjusting the mesh set
set mesh=inactive.
2.2.2Changing the background model¶
Adjustable parameters: model type, v0
To change the value of the constant background velocity model:
- Set
model type=background. This will change the view to the background modelv0. - Adjust
v0using the slider. - When finished ajusting set
model type=block
2.2.3Changing the parameters of a single block¶
Adjustable parameters: add block, model type, v1, xc, zc, dx, dz
To change the parameters of a single block model (default):
- Set
add block=active. - Adjust
v1,xc,zc,dx,dzusing the sliders. The original block settings will remain in view. - When finished ajusting set
model type=background - To confirm the parameter changes and veiw the current block, set
model type=block
2.2.4Adding more blocks¶
Adjustable parameters: add block, v1, xc, zc, dx, dz
The model can contain multiple blocks. To add new blocks to the model:
- Set
add block=inactive - Adjust the parameters of the new block
v1,xc,zc,dx,dz. The velocity model will not change yet. The white boundary of the new block serves as a preview. - When finished adjusting the new block parameters set
add block=active. The velocity model will be updated with the new block. - Repeat Steps 1-3 to add more blocks.
The parameters of the blocks in a multi-block model cannot be changed once the block has been added. Attempting to adjust the parameters after the block has been added to the model (following the directions in the previous section “Changing the parameters of a single block”) will remove all previous blocks. A single block model with the current block is created.
app = TomographyInversionApp()
app.interact_plot_model()2.3Step 2: Simulate travel time data and add noise¶
Within this app, by using the velocity model set up above, we simulate traveltime tomography data and add Gaussian noise. This syntehtic data set will be used in the following inversion module. Controlling parameters are:
percent (%): percentage of the Gaussian noisefloor (s): floor of the Gaussain noiserandom seed: seed to generate random variables having normal distributiontx_rx_planeorhistogram: data display optionsupdate: this buttion is for the interaction with the Step 1 app. If the velocity model is changed by altering the first app, you can simply clickupdateto run simulation again with the updated velocity model.
app.interact_data()2.4Step 3: inversion¶
Here we invert the data generated in Step 2 and explore the results. Step 3 is divided into four sections:
- Step 3.1: Run inversion
- Step 3.2: Plot recovered model
- Step 3.3: Plot predicted data
- Step 3.4: Plot misfit curves
2.4.1Step 3.1: Run inversion¶
The inversion, run in the cell below, requires the following input parameters, each of which can be adjusted:
maxIter: maximum number of iterationsm0: initial modelmref: reference modelpercentage: percent standard deviation for the uncertaintyfloor: floor value for the uncertaintychifact: chifactor for the target misfitbeta0_ratio: ratio to set the initial betacoolingFactor: cooling factor to cool betan_iter_per_beta: # of interations for each beta valuealpha_s:alpha_x:alpha_z:use_target: use target misfit as a stopping criteria or not
model, pred, save = app.run_inversion(
maxIter=20,
m0=1./1000.,
mref=1./1000.,
percentage=0,
floor=0.01,
chifact=0.1,
beta0_ratio=1e2,
coolingFactor=2,
n_iter_per_beta=1,
alpha_s=1/(app.mesh_prop.hx.min())**2,
alpha_x=1,
alpha_z=1,
use_target=True
)0.1
SimPEG.InvProblem is setting bfgsH0 to the inverse of the eval2Deriv.
***Done using same Solver and solverOpts as the problem***
SimPEG.SaveOutputEveryIteration will save your inversion progress as: '###-InversionModel-2021-03-11-21-26.txt'
model has any nan: 0
=============================== Projected GNCG ===============================
# beta phi_d phi_m f |proj(x-g)-x| LS Comment
-----------------------------------------------------------------------------
x0 has any nan: 0
0 1.59e+07 3.40e+02 0.00e+00 3.40e+02 1.12e+04 0
1 7.97e+06 1.47e+02 1.07e-06 1.56e+02 4.92e+04 0
2 3.98e+06 1.33e+02 2.36e-06 1.42e+02 4.72e+04 0 Skip BFGS
3 1.99e+06 1.16e+02 5.55e-06 1.27e+02 4.31e+04 0 Skip BFGS
4 9.96e+05 9.70e+01 1.23e-05 1.09e+02 3.79e+04 0 Skip BFGS
5 4.98e+05 7.91e+01 2.51e-05 9.17e+01 3.24e+04 0 Skip BFGS
6 2.49e+05 6.27e+01 4.89e-05 7.49e+01 2.67e+04 0 Skip BFGS
7 1.24e+05 4.85e+01 8.96e-05 5.96e+01 2.10e+04 0 Skip BFGS
8 6.22e+04 3.73e+01 1.53e-04 4.68e+01 1.61e+04 0 Skip BFGS
9 3.11e+04 2.87e+01 2.52e-04 3.65e+01 1.24e+04 0 Skip BFGS
10 1.56e+04 2.14e+01 4.18e-04 2.79e+01 9.50e+03 0 Skip BFGS
11 7.78e+03 1.51e+01 7.09e-04 2.06e+01 7.05e+03 0 Skip BFGS
12 3.89e+03 1.00e+01 1.17e-03 1.46e+01 5.21e+03 0 Skip BFGS
13 1.94e+03 6.53e+00 1.80e-03 1.00e+01 3.32e+03 0 Skip BFGS
------------------------- STOP! -------------------------
1 : |fc-fOld| = 0.0000e+00 <= tolF*(1+|f0|) = 3.4119e+01
1 : |xc-x_last| = 3.0710e-04 <= tolX*(1+|x0|) = 1.0141e-01
0 : |proj(x-g)-x| = 3.3180e+03 <= tolG = 1.0000e-10
0 : |proj(x-g)-x| = 3.3180e+03 <= 1e3*eps = 1.0000e-07
0 : maxIter = 20 <= iter = 14
------------------------- DONE! -------------------------
2.4.2Step 3.2: Plot recovered model¶
The app below displays the:
- Recovered model from the inversion in Step 3.1 (left)
- True model generated in Step 1 (right)
The model view is changed using:
ii: each iteration of the inversion (ii=0 is the initial modelm0)fixed: when selected the colorbar of both plots have the same fixed limits
app.interact_model_inversion(model, clim=None)2.4.3Step 3.3: Plot predicted data¶
The app below displays the:
- Obversed data calculated in Step 2 (left)
- Predicted data from the inversion in Step 3.1 (center)
- Normalized misfit between the observed and predicted data (right)
The predicted data and normalized misfit view is changed using:
ii: each iteration of the inversion (ii=0 is the initial modelm0)fixed: when selected the colorbar of both data plots have the same fixed limits
app.interact_data_inversion(pred)2.4.4Step 3.4: Plot misfit curves¶
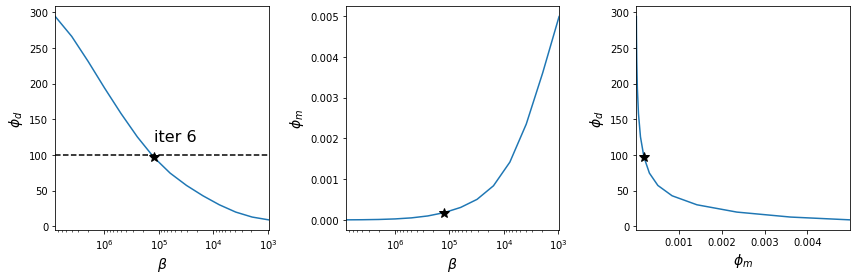
- Plot of the data misfit and model norm values as functions of inversion iteration
- Tikhonov curves with the optimal values indicated by the star
- Data misfit as a function of trade-off parameter β
- Model norm as a function of trade-off parameter β
- Data misfit versus model norm
app.plot_tikhonov_curves(save);